Imaging system finds new drug models
By C. G. Masi, Contributing Editor
Over the past few years, pharmaceutical companies have made impressive strides in automating the analysis and characterization of important biological molecules used to develop pharmaceutical compounds. Scientists are synthesizing novel compounds that can effectively bind to specific protein targets and disrupt the biochemical process of a devastating disease. However, this process is a long and complex one and requires astronomical numbers of experiments. What makes this approach possible is the implementation of automated networks, actions, and events throughout the experimental process. Part of that automation involves a video microscope system that takes images of millions of individual protein crystals and then automatically screens them for size, shape, and growth.
For example, Gene Logic (Gaithersburg, MD) provides devices that sort through the biochemical pathways involved in a specific disease and identify proteins that are potentially high-quality targets of opportunity for chemical intervention. Then, a biotechnology company, such as Syrrx (San Diego, CA), uses the information to devise synthetic compounds that will bind to the target proteins and disrupt a diseased process.
"Syrrx is a biotech company working its way toward becoming a pharmaceutical company," explains Keith P. Wilson, vice president of technology. "We can capture actual pictures of the proteins that a particular drug might interact with and use that information to guide compound design." These pictures are crystallographic images that help crystallographers to determine a protein crystal's structure (see Fig. 1, below).
FIGURE 1. Currently, x-ray crystallography is the most effective tool for elucidating the three-dimensional structures of proteins. To create an image, the crystallographer places a single protein crystal in a monochromatic x-ray beam. Each dark spot represents a reflection from a set of parallel crystal planes. Based on x-ray diffraction theory, experienced scientists work backward from the arrangement of spots on the image to the three-dimensional arrangement of atoms in the protein molecules.
The difficult part of the procedure is getting the proteins to crystallize in the first place, so that crystallographers can determine their three-dimensional structures. "We have a high-throughput crystallization system," says Ken Goodwill, Syrrx director of project management. "We try and examine as many different parameters as possible, because there is no general rule of thumb for how to crystallize proteins. Each protein is essentially its own science project."
Therefore, the procedure involves thousands of small crystallization experiments for each protein target. Each experiment attempts to grow crystals under slightly different conditions. Most of these experiments don't work, some work a little, and others work successfully.
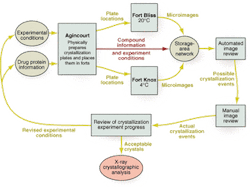
The major problem is visualizing the crystals as they grow. "You must have a microscope system," says Brian Karlak, Syrrx director of informatics, "to take pictures of these nanodrops, which are on the order of microns. In addition, you need a system to go through the millions of produced images to find those that have crystals in them." Each well in a holding plate contains an individual crystallization experiment, and each experiment incubates for up to a month in one of two huge storage vaults called 'forts' (see Fig. 2). We call them Fort Bliss, which is kept at 20°C, and Fort Knox, which is maintained at 4°C," says Karlak.
FIGURE 2. Syrrx runs its crystallization experiments on 96-well plates. The crystals grow in small droplets, which appear as white circles in black squares. The black squares are actually small chambers. The droplets are in diffusive contact with the larger volume of fluid in the remainder of the well (larger square). Giving the larger fluid volume, a higher salinity than the droplet sets up a thermodynamic gradient that draws water from the droplet to the larger volume. The droplet slowly shrinks and concentrates the dissolved protein molecules. Sometimes, this drying process induces the proteins to form a crystal.
Each fort can store as many as 10,000 plates. With 96 experiments running in each plate, Syrrx has the capacity to run up to 1.92 million experiments simultaneously. If each experiment runs for a month, the system's throughput is more than 23 million experiments per year.
null
A robotic imaging system images each plate on schedules of 6 hours, 24 hours, 72 hours, 7 days, 14 days, and 28 days after it goes into its fort (see Fig. 3). This process results in the collection of six images of each well on each plate in each fort, or some 11.5 million images per year. All those images, along with data on the experimental conditions for each well, are stored in a database on a terabyte-sized storage-area network (SAN).
Imaging-system operation begins with an initial set of experimental conditions that is used with each protein or drug/protein complex to be crystallized. The robot, which is named Agincourt, uses commands supplied by the Syrrx synthetic chemists (see Fig. 4). It assembles the experiments in each well of each 96-well plate, seals the plate with clear tape, and stores the plates in the forts.
FIGURE 4. Protein crystallization process begins with an initial set of experimental conditions. A robot assembles the experiments in each well of each 96-well plate, seals the plates, and stores them in temperature-controlled storage areas called Fort Bliss and Fort Knox. Video microscopes in the forts image each crystallization experiment individually, searching for growing crystals. The process ends when one or more crystals appear with a size and quality to provide for analysis by a crystallographer.
Crystal images
Each fort contains, in addition to locations to store crystallization plates, two video microscopes with automated stages and a robot to move plates between the microscope stages and their storage locations. Since the standard imaging schedule for each plate is not the same-the interval between images increases as the plates get older-and each fort contains plates of widely varying ages, there is no simple program for selecting which and when to image each plate. The plate-handling robot must, every time one of the video microscopes finishes imaging a plate, query the SAN over the communications network about which plates are due for an image.
null
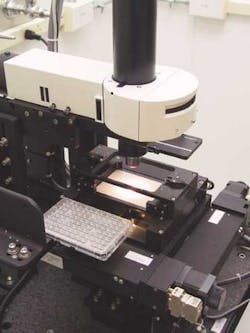
The crystals, if they grow at all, are very small; for example, one droplet might be growing as many as five protein crystals. The imaging station consists of a computer-controlled stage, a video microscope, and an image-acquisition computer (see photo above). The lens used in the video microscope is an Olympus America (Melville, NY) MPlan 5x /0.10, which does not include autofocus. Because all the plates are made to exacting tolerances and the crystallization droplets are very small, there is no need to refocus the lens for each image, which saves process time.
The camera is a Dalstar 1M15 from Dalsa (Waterloo, ON, Canada). It provides 1024 × 1024-pixel resolution at 15 frames/s, a 20-MHz data rate, and a 12-bit, RS-422 data format. During system operation, the imaging station's dual- Pentium III host computer automatically crops each black-and-white image to 700 × 750 pixels before sending it to the image-acquisition and storage computer.
The acquired images are delivered over the system's Gigabit-Ethernet link to a 2-node cluster computer. This cluster acquires and manages the Syrrx image database, which is stored on a 2-Tbyte SAN. The SAN consists of a set of Hewlett-Packard (Littleton, MA) ProLiant DL580 servers, each containing three Pentium III CPUs and 16 Gbytes of RAM. The Linux-based cluster communicates commands and header data over the Gigabit-Ethernet link, and images are sent over a dedicated high-speed optical-fiber network.
To analyze the images, Syrrx resorts to its 250-node-cluster supercomputer and a 16-Tbyte SAN. This large SAN consists of IBM Corp. (Bethesda, MD) X330 servers that run dual Pentium III CPUs and move data over another high-speed optical-fiber network.
All these systems communicate with each other and with the scientists who ultimately use the data via the Gigabit-Ethernet link. Because the image analysis is a data-intensive computational exercise, the cluster computers operate on their own internal optical fiber network specially optimized to move image data between the nodes and the cluster's SAN. This setup keeps the computers from bogging down under the huge data communications load.
Small odds
"A key point," Goodwill says, "is that the overall success rate for these experiments is approximately one in a thousand. So if we run, say, 50,000 crystallization experiments in a day, there will be maybe 50 that we need to look at closely. The key is to define those 50."
Part of the robotic-based imaging system job is to sweep away all the obvious null data and flag every well that shows a hint of crystallization. "This automatic operating system determines whether or not there is a crystal based on an algorithm developed by Strand Genomics (Columbia, MD)," Karlak says. "This algorithm identifies features, such as edges, lines, and contrasts, associated with crystal images."
Strand Genomics devised a five-way classifier to extract significant features. The algorithm, Sphatika, has demonstrated an accuracy of more than 90%. Once the algorithm sees features that lead it to believe a crystal is growing, it flags the image for review by a scientist. It then proceeds to estimate the crystal's size and shape. This information goes into an electronic report for the scientist responsible for that batch of experiments. Using a workstation, the scientist manually calls each flagged image for review and then decides what to do next. Most often, the next step is to run another series of experiments with modified parameters based on the most successful results from the last iteration. However, if the scientist is satisfied that this last crystal is satisfactory, the crystallization effort is halted and the best crystals are sent on for x-ray analysis.
Although this method of drug discovery is new (Syrrx was started in 1999), it has produced, to date, five promising anticancer drugs, two of which have already received US Food and Drug Administration approval.