Blind deconvolution help 3-D reconstruction of microscope images
Blind deconvolution help 3-D reconstruction of microscope images
By Lawrence J. Curran, Contributing Editor
Three-dimensional (3-D) microscopy is widely used in medical applications and biosciences, but the images obtained are distorted by both the specimen under study and the optics of the imaging system. Confocal microscopy eliminates some of the distortion, but confocal microscopes are expensive and often difficult to use. In contrast, an image-processing technique called deconvolution is an effective tool that is used to reduce blurring.
Moreover, AutoDeblur software, developed by AutoQuant Imaging Inc. (Watervliet, NY), can largely reverse most of the distortion in microscopic imaging by means of a process called blind deconvolution. AutoDeblur applies 3-D blind-deconvolution algorithms that automatically remove haze, blur, noise, and other artifacts from 3-D micrographs.
In operation, blind deconvolution derives many different PSF values as it images 3-D specimens and applies what is essentially a 3-D fast-Fourier-transform (FFT) algorithm to each point-speed function (PSF) in an iterative process that reverses or corrects for virtually all of the distortion. It is especially effective in visualizing the 3-D data associated with thick microscopic samples, improving the sensitivity and axial resolution of a microscope and allowing users to better detect fine structures and perform more accurate morphology.
The software`s iterative blind-deconvolution technology also allows users to apply deconvolution techniques without going through the tedious, esoteric, and error-prone procedure of measuring the PSF of the microscope in use. "AutoDeblur does that automatically in a method that`s also inherently adaptive, yielding results that are better than those obtainable with other deconvolution methods," says David Hitrys, a spokesman for AutoQuant Imaging and president of Cell Dynamics Inc. (Framingham, MA), an integrator of multidimensional microscopy systems.
Cell Dynamics has the exclusive worldwide license to sell AutoDeblur, and Hitrys has helped to develop the software`s user interface. He points out that blind deconvolution is better than regular deconvolution methods because it adapts to changes in the PSF caused by variations in the refractive index and the microscope`s optics. "A biological specimen consists of a 3-D volume, and the blind-deconvolution algorithm breaks that volume into several cubes or 3-D chunks," explains Hitrys. "Each cube undergoes a 3-D FFT that shows the spatial frequencies in that little subvolume, all of which are used to reverse distortions," he adds. Blind deconvolution therefore results in much clearer images than those based on a single PSF.
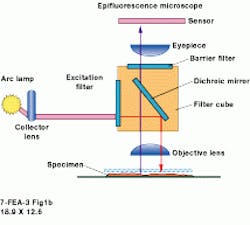
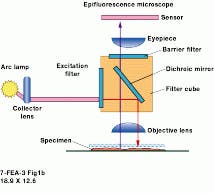
At the University of Connecticut (UConn; Storrs, CT) Biotechnology Center Imaging Applications Facility they are getting good results imaging insect eggs in a DNA study being conducted there. Hal Krider, professor of molecular and cell biology and head of the facility, says that with AutoDeblur his students can improve almost any image. "It provides a ready and straightforward solution to deblurring images quickly," he adds. At UConn, the imaging application program runs on a 200-MHz Pentium Pro PC under Windows NT, but it can be used on any Windows-Intel computer, as well as on a Unix machine, without the need for additional hardware (see Fig. 1).
Timothy Holmes, president of AutoQuant Imaging, says AutoDeblur is based on more than a decade of research. "This is the most advanced 3-D blind-deconvolution product currently available," he contends. "It incorporates a variety of algorithms ranging from simple to quick to sophisticated, all of which are supported by an easy-to-use graphical interface" (see "Algorithms do it blindly," p. 32). In addition, AutoDeblur can be used to perform 3-D deconvolution for several forms of microscopy: transmitted-light brightfield, widefield epifluorescence, and confocal pinhole laser-scanned epifluorescence micro scopy (see "Deconvolution glossary" p. 31).
Holmes explains that for a confocal fluorescence microscope, images of adjacent planes are sequentially digitized and optically sectioned in a manner similar to the widefield case, except that each frame is collected by using a raster-scanned laser spot and a photomultiplier pinhole detector. Each confocal optical section is, by itself, already relatively deblurred because the confocal optics reject most of the out-of-focus light.
Confocal compromise
Despite this improvement over a widefield microscope, however, the confocal microscope has some limitations. While it rejects most of the out-of-focus light, the confocal microscope doesn`t reject all of it, thereby retaining an obvious haze--albeit a haze that is greatly reduced compared to results using a widefield microscope.
Because of this light rejection, Holmes says, far fewer photons are detected, possibly introducing a substantial quantum-photon noise component into the raw data. The noise routinely causes limitations in protocols where very fine structures need to be seen. For example, it may be difficult to determine whether a void in the image intensity is caused by a fine structure that is really there, or if it results from erroneous, random fluctuations caused by the statistical nature of quantum noise.
In confocal fluorescence microscopy, AutoDeblur`s deblurring algorithms reduce these undesirable effects, enhancing the utility of the microscope. AutoDeblur is also valuable in conventional wide-field microscopy. According to Hitrys, it captures much more data about the specimen than can be obtained with a confocal microscope. He says that more data capture takes place because in rejecting out-of-focus light, a confocal microscope can eliminate some valuable information about the specimen. AutoDeblur extracts the information contained in the haze gathered in images from a wide-field microscope, reverses the haze, and uncovers valuable information in the specimen faster than is possible with a confocal microscope, claims Hitrys.
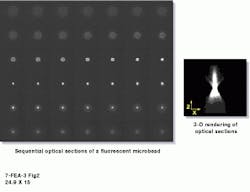
He also points out that a confocal microscope sells for as much as $180,000, adding that deconvolution technology was originally described as "a poor man`s confocal microscope." But because of advancements in the price-performance ratios of PCs and high-performance charge-coupled-device (CCD) cameras, "some of today`s most exciting work is being done with wide-field microscopy coupled with deconvolution software ," states Hitrys (see Fig. 2). He adds that a typical system configuration--consisting of a wide-field microscope, camera, stepper motor, computer, and software--sells for about $80,000.
Such a system approximates the core combination for blind deconvolution that Krider uses at the UConn facility. "We wind up with every kind of imaging experience and problem you can imagine," Krider says. The facility also supports course work at UConn and conducts funded research for outside clients. It usually has six or eight imaging projects in progress at any time. Applications range from studies of how cell changes affect PH levels to helping a food company determine why some of its cookies stick to the package.
Furthermore, Krider`s facility is using the blind-deconvolution technique embodied in AutoDeblur for a study of DNA replication in the eggs of fruit flies. "It is an organism much appreciated by the scientific community," notes Krider. This is because the fruit fly is domesticated for use in genetics laboratories and is "profoundly informative" in understanding the early development of organisms, Krider points out.
He explains that all cells replicate their DNA as they develop, "which is usually a straightforward process, but in special cases, some genes are excessively over-replicated. This can happen in all organisms, and, while it isn`t always a problem, this over-replication can lead to cancer and other abnormalities in humans, so we want to understand the process," says Krider.
As part of nature, a group of cells around the egg of a fruit fly quickly wraps the egg in a "shell" of protein. In creating that protective shell, some genes become massively over-replicated. By applying blind deconvolution on cell images, the genes can readily be seen as lumps.
"We study this because there are mutations in genes that we think can regulate or interfere with the process--a group of mutations--that identify the genes that are important DNA information carriers," Krider explains. "We want to see how they affect this usually massive over-replication. We`re looking for things that control, or fail to control, this unusual replication," he says. Understanding this process can add to the knowledge of how cancer and other abnormal cells replicate.
Krider points out that a confocal microscope may be the ideal instrument to use in these imaging experiments because it eliminates some of the haze or blur that results in 3-D sections. The UConn facility has one, "but a confocal microscope is an expensive and demanding instrument that usually requires substantial learning before it can be used effectively," he says. "It`s not a casual imaging device, because using it requires a lot of preparation of the material to be imaged. It is usually the last step in an experiment," remarks Krider.
Therefore, Krider bought a high-resolution camera and added AutoDeblur to a high-performance PC that serves as the "main deconvolution processor." This system essentially removes all of the blurring captured during imaging. The UConn facility uses eight other microscopes. Each micro scope is tied to a networked computer, and each microscope can be used by students more casually than is possible with a confocal microscope to obtain images of fruit-fly eggs.
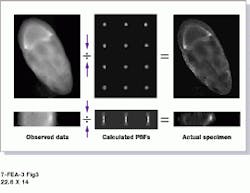
The main deconvolution processor uses a 200-MHz Pentium Pro running Windows NT and AutoDeblur. This system allows UConn researchers to isolate the blobs created during the massive over-replication process, yielding a blur-free image after several iterations of the blind deconvolution algorithms (see Fig. 3). "We bought a high-resolution camera and Auto Deblur so that the students could concentrate on biology, get the best possible images with the conventional microscopes they normally use, then run AutoDeblur later on the deconvolution processor," Krider explains. "We don`t have to use a confocal microscope, but we need one fast computer," he adds.
The high-resolution camera, an Olympix Peltier-cooled CCD digital camera from the Precision Instrument Division of Olympus America Inc. (Melville, NY), provides a 1317 ¥ 1035-pixel format, a 12-bit depth, and an 8-MHz interface. The camera is equipped with software drivers that direct it to take stepped, timed images, and to transport them in several formats, including TIFF (tagged image file format).
Another helpful software package in the UConn facility is Scion Image from Scion Corp. (Frederick, MD), which complements AutoDeblur by displaying and reading the stack of images that AutoDeblur creates. "It`s public domain software that worked the first time we used it," Krider reports.
Although the UConn facility is still in the early stages of working with both software packages, blind deconvolution has made several contributions to its research. It eliminates a large amount of repetitive preparation, such as the fixation of a specimen. "We can often keep the protein around, but we can`t always keep the fixation," says Krider. This means steps have to be repeated to bring the material to a state in which it will work in an experiment.
What`s more, use of blind deconvolution yields clearer images earlier than are possible with other methods. "As features emerge in any of the images we obtain, I can send them to the deconvolution processor and ask it what those features look like," Krider notes.
FIGURE 1. In an automated microscopy system at the University of Connecticut Biotechnology Center Imaging Applications Facility, a core deconvolution processor contains a Pentium-based personal computer that runs under Windows 95/98/NT and is coupled to a widefield microscope and a high-resolution CCD camera.
FIGURE 2. With AutoBlur software, blind convolution can be shown as a two-dimensional representation of multiple point-spread functions (PSFs). A series of through-focus microscopic images of a 200-nm microbead illustrates a measured PSF (left). A three-dimensional rendering of several PSFs is used to reverse distortions in the microscopic images and to compile an x-y (orthogonal) view (right).
FIGURE 3. In a typical deconvolution procedure using AutoDeblur, the observed-data image (left) is processed using the blind deconvolution algorithm in an iterative process that calculates multiple PSF values (center). Ultimately, the processing delivers a deblurred image (right).
Deconvolution glossary
Confocal microscopy--an optical-physical technique that increases the spatial resolution to reduce the depth of field of a microscope. The technique is used to reject out-of-focus signals that cause scattering, but information in those signals can be helpful.
Epifluorescence microscopy--a method of illuminating fluorophosphors in a manner that allows effective lighting of a target specimen and the collection of its emissions.
Fluorescence--the use of a fluorophosphor to chemically tag a specimen of interest with fluorescent properties to enhance its visibility.
Morphology--the study of changes in the shape, size, or volume of cells.
Transmitted-light brightfield microscopy--wide-field microscopy that uses white light rather than fluorescence to illuminate a target, which absorbs the light, yielding contrast.
Wide-field microscopy--a general term that refers to the use of a normal, conventional microscope. The technique has become more attractive because, in combination with deconvolution, it offers an economical tool for use in the biosciences.
Algorithms do it blindly
L. C.
Deconvolution algorithms work with both optically sectioned 3-D wide-field imagery and with 3-D confocal fluorescence imagery. Some algorithms add an effective signal-to-noise improvement in very-low-light-level conditions, effectively improving the low-light sensitivity of the microscope.
None of the algorithms in AutoDeblur require a point-spread- function (PSF) measurement. The blind-deconvolution algorithms automatically produce a reconstruction of the PSF from the noise image data.
Blind deconvolution is an established, robust algorithm, but is least often used because it is slow. However, its superior performance against noise is needed in rare cases--for instance, those that have extremely severe signal-to-noise levels (10 photons per pixel or fewer).
The power-accelerated and turbo-speed blind deconvolutions are the two most-often-used algorithms because of their superior speed. These are still robust enough against noise, even at the high-speed settings.
Any combination of a wide-field microscope and a digitizing camera will contain nonuniformities in its flat-field response. These nonuniformities might be caused by dust in the optics or flaws in photodetector response. There also may be erroneous black levels (dark currents) and other problems with photodetectors. To perform deconvolution, it is important to correct these common problems.
Some image-processing software used in microscopy can correct for these errors. However, this software usually isn`t designed to work with deconvolution, and the corrections aren`t generally compatible with effective deconvolution. AutoDeblur is specifically designed to correct for such errors in a manner that supports effective deconvolution.
L. C.